SPInDel kit
The Species Identification by Insertions/Deletions (SPInDel) kit allow the identification of human genetic material on a biological sample, a preliminary step in many investigations. The kit is an alternative to DNA sequencing that uses a conventional genotyping methodology similar to that employed with Short Tandem Repeats (STRs), involving multiplex PCR followed by fragment size determination using capillary electrophoresis.
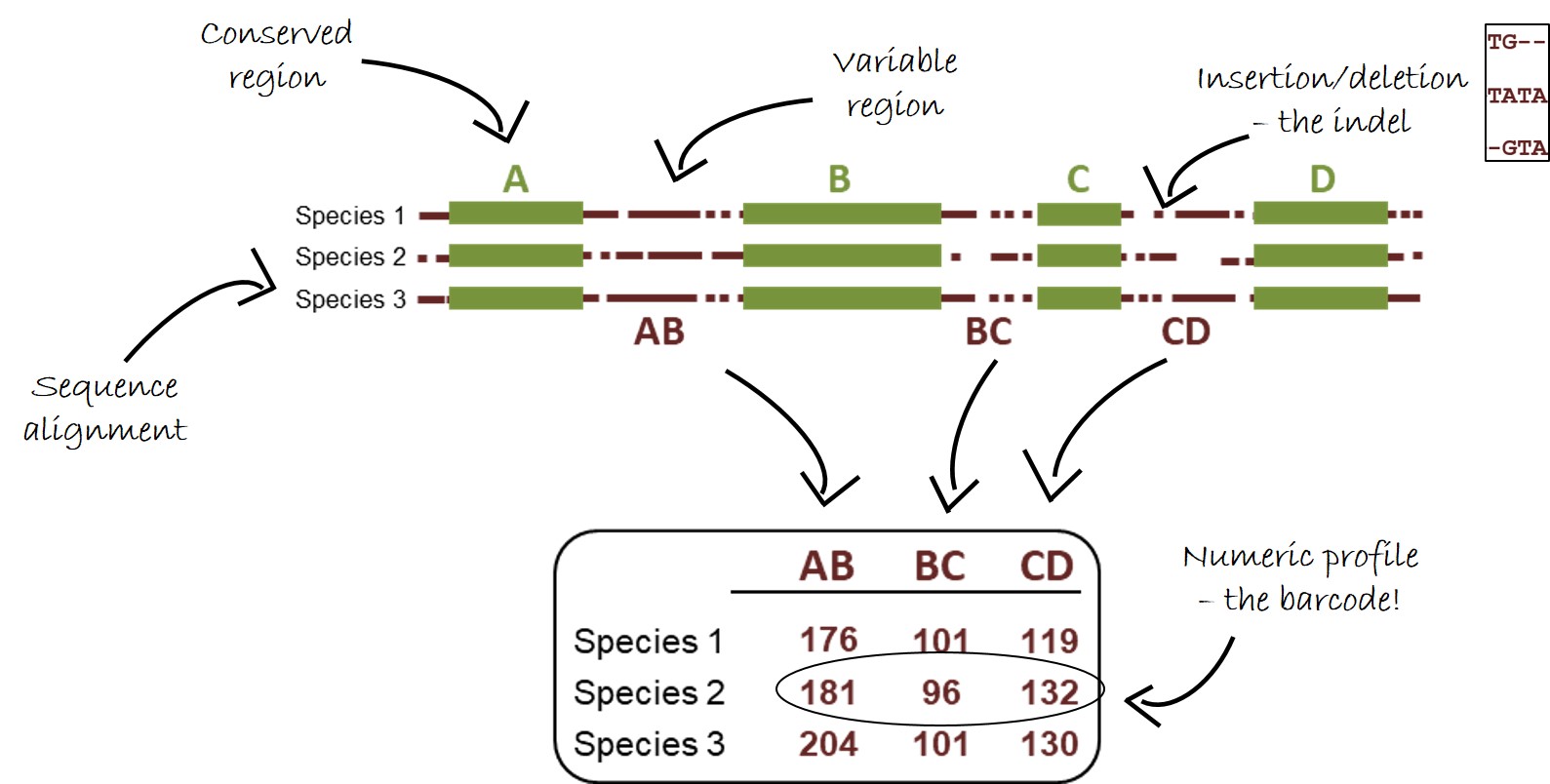
The identification of species with the SPInDel method is achieved by the amplification of six hypervariable regions in mitochondrial ribosomal RNA (rRNA) genes using highly conserved PCR primers. Each species is defined by a unique numeric profile of fragment lengths (i.e., a numeric barcode) resulting from the combination of the length of indel-rich regions.
The kit identifies several mammalian species, including Cat (Felis catus), Dog (Canis familiaris), Cattle (Bos taurus), Goat (Capra hircus), Pig (Sus scrofa), Horse (Equus caballus), Rabbit (Oryctolagus cuniculus), Sheep (Ovis aries) and Mouse (Mus musculus). Several wildlife species are also identified (see complete list below).
The kit includes an allelic ladder to help in the analyses.
The identification of the species can be done using the SPIndel Kit Profile Identification webpage.
Specifications and recommendations(others setting may be used with proper adaptations) |
|
| Components | SPInDel Primer Set (10x) SPInDel Allelic Ladder |
| Product Size | 50 reactions (K001AL) 100 reactions (K001BL) |
| Equipment | ABI Genetic Analyzer (310, 3100, 3130, 3500, 3730 or equivalent) |
| Dye | 6-FAM |
| Size Standard | GeneScan™ 500 LIZ® |
| Filter Set | G5 |
| Separation Polymer | POP7, POP6, POP4 |
| Sample Type | DNA |
| Sample Volume | 10 µL/reaction |
| Technique | Fragment size determination (similar to STRs) |
| Shipping Condition | Dry, room temperature |
THE CONCEPT
The identification of species with the SPInDel method is achieved by the amplification of six hypervariable regions in mitochondrial ribosomal RNA (rRNA) genes using highly conserved PCR primers. Each species is defined by a unique numeric profile of fragment lengths (i.e., a numeric barcode) resulting from the combination of the length of indel-rich regions.
HOW IT WORKS
The kit is an alternative to DNA sequencing that uses a conventional genotyping methodology similar to that employed with Short Tandem Repeats (STRs), involving multiplex PCR followed by fragment size determination using capillary electrophoresis. The kit includes a software to help in the data analyses.
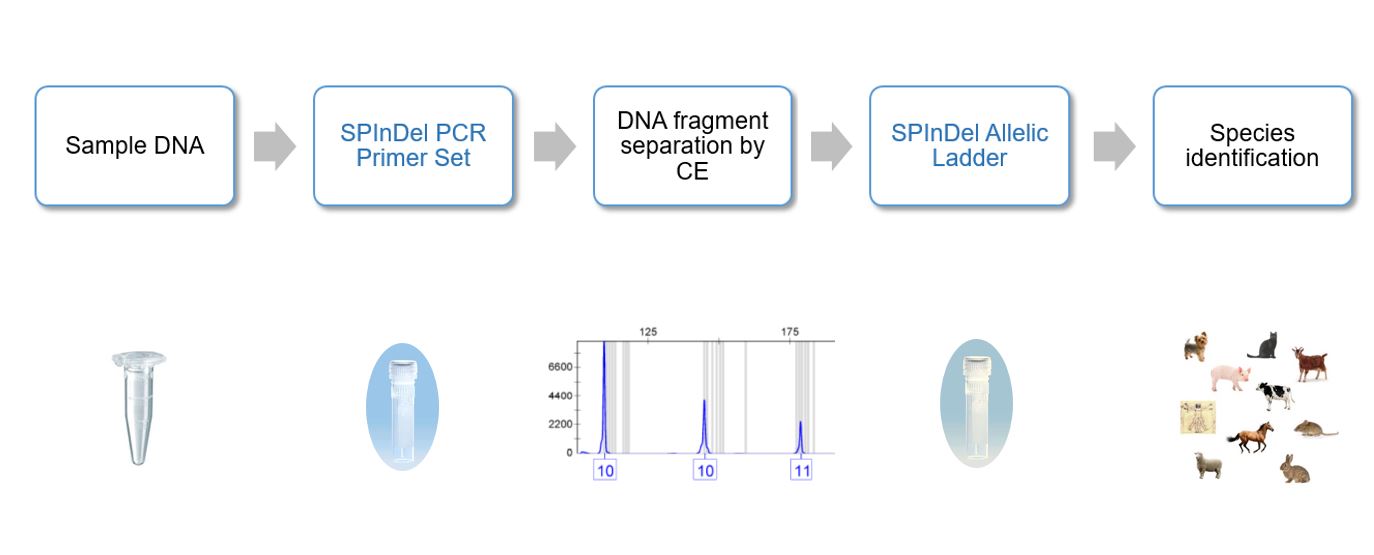
SPInDel PROFILE
TYPES OF SAMPLES SUCCESSFULLY TESTED

ADVANTAGES
APPROPRIATE FOR LOW-QUANTITY AND/OR DEGRADED DNA SAMPLES
The SPInDel targets the mitochondrial DNA (mtDNA), which is present in many copies per cell, and amplifies short DNA fragments (3 out of 6 loci have less than 200 bp). Therefore, it is appropriate for difficult samples with degraded DNA, where nuclear DNA-based methods fail.
SUITABLE FOR DETECTION OF MIXTURES
The SPInDel assay uses fragments lengths to achieve identifications. Because different species have different fragment lengths, they are easy discriminated in a sample. This feature overcomes one important drawback of DNA sequencing-based methods.
APPROPRIATE FOR A HIGH THROUGHPUT SAMPLE ANALYSIS
The use of less time-consuming, cost-effective and efficient automated fluorescent DNA detection makes our method suitable for high throughput sample analysis with conventional laboratory equipment.
INCREASED GENOTYPING EFFICIENCY
The simultaneous amplification of 6 mtDNA regions considerably increases the discriminatory efficiency of the procedure by avoiding the complete absence of results in cases with non-amplified loci – a severe limitation of methods relying on singleplex PCR.
NO NEED FOR DNA SEQUENCING
The SPInDel assay only requires an electrophoresis after PCR. There is no need for DNA sequencing reactions, making our method less time-consuming and expensive.
SUITABLE FOR DETECTION OF A WIDE RANGE OF SPECIES
The design of PCR primers in highly conserved mtDNA regions is particularly useful for a broad range of species detection. Successful discriminations are achieved among several species of mammals, birds and fishes.
SPECIES WITH UNIQUE SPInDel PROFILES
Homo sapiens (Human)
Bos taurus (Cattle)
Canis lupus (Dog/Wolf)
Capra hircus (Goat)
Equus caballus (Horse)
Felis catus (Cat)
Mus musculus (Mouse)
Oryctolagus cuniculus (Rabbit)
Ovis aries (Sheep)
Sus scrofa (Domestic pig/Wild boar)
Capra pyrenaica (Iberian ibex)
Rattus norvegicus (Brown rat)
Martes foina (Beech marten)
Mustela putorius furo (Ferret)
Meles meles (European badger)
Nyctereutes procyonides (Racoon dog)
Atelerix albiventris (African pygmy hedgehog)
Mesocricetus auratus (Hamster)
Cavia porcellus (Guinea pig)
Genetta genetta (Common genet)
Cervus elaphus (Red deer)
Capreolus capreolus (Roe deer)
Dama dama (Fallow deer)
Alces alces (Moose)
Ursus arctos (Brown bear)
Equus asinus (Donkey) / Equus africanus (African wild ass)
Vulpes vulpes (Red fox)
Vicugna pacos (Alpaca)
Sarcophilus harrisii (Tasmanian devil)
Dasyurus viverrinus (Eastern quoll)
Dasyurus maculatus (Spotted-tail quoll)
Trichosurus vulpecula (Brushtail possum)
Thylogale billardierii (Tasmanian pademelon)
Macropus rufogriseus rufogriseus (Red-necked wallaby)
Potorous tridactylus (Long-nosed potoroo)
PUBLICATIONS
Pereira F, Carneiro J, Matthiesen R, van Asch B, Pinto N, Gusmão L, Amorim A.
Identification of species by multiplex analysis of variable-length sequences.
Nucleic Acids Research. 2010. 38 (22): e203.
Carneiro J, Pereira F, Amorim A.
SPInDel: a multi-functional workbench for species identification using insertion/deletion variants.
Molecular Ecology Resources. 2012. 12(6):1190-5.
Gonçalves J, Marks CA, Obendorf D, Amorim A, Pereira F.
A multiplex PCR assay for identification of the red fox (Vulpes vulpes) using the mitochondrial ribosomal RNA genes
Conservation Genetics Resources. 2015. 7(1): 45-48. DOI: 10.1007/s12686-014-0343-0
Alves C, Pereira R, Prieto L, Aler M, Amaral C, Arévalo C, Berardi G, Di Rocco F, Caputo M, Carmona CH, Catelli L, Costal HA, Coufalova P, Furfuro S, García O, Gaviria A, Goios A, Gómez JJB, Hernández A, Hernández ECB, Miranda L, Parra D, Pedrosa S, Porto MJA, Rebelo ML, Spirito M, Torres MCV, Amorim A, Pereira F.
Species identification in forensic samples using the SPInDel approach: A GHEP-ISFG inter-laboratory collaborative exercise
Forensic Science International: Genetics. 2017. 28: 219–224. DOI: http://dx.doi.org/10.1016/j.fsigen.2017.03.003
Pereira F, Alves C, Couto C, Díaz LL, Parra D, Furfuro S, Aler M, Borrego LB, Olekšáková T, Balsa F, Sampaio L, Porto MJA, Costa HA, Voss CA, Caputo M, Corach D, García O, Moro SP, Pereira R, Amorim A
Species identification in routine casework samples using the SPInDel kit.
Forensic Science International: Genetics Supplement Series. 2019. DOI: 10.1016/j.fsigss.2019.09.070